More VentureScript: Linear Regression
In this chapter, we will build up a somewhat more elaborate model, and
explore strategies for inspecting and debugging it. The problem we
address is linear regression: trying to infer a linear relationship
between an input and an output from some data. This is of course
well-trodden terrain -- we are primarily using it to introduce more
VentureScript concepts, but the models themselves will get interesting
toward the end.
Contents
Recap
In the previous part we discussed basic
concepts of VentureScript programming, modeling, and inference. As a
reminder, the main modes of using VentureScript we saw are starting Venture
interactively
$ venture
and running standalone VentureScript scripts
$ venture -f script.vnts
Simple Linear Regression
Let's start by putting down the first model for linear regression that
comes to mind. A line has a slope and an intercept
assume intercept = normal(0,10);
assume slope = normal(0,10)
10.4725503978
and is reasonable to represent as a function from the independent to
the dependent variable
assume line = proc(x) { slope * x + intercept }
<procedure>
In VentureScript, proc is the syntax for making an anonymous
function with the given formal parameters and behavior, and we use
assume to give it a name in our model.
Finally, we will model our data observations as subject to Gaussian
noise, so we capture that by defining an observation function
assume obs = proc(x) { normal(line(x), 1) }
<procedure>
If you don't want to type that, you can download
lin-reg-1.vnts and use it with
$ venture -f lin-reg-1.vnts --interactive
Let's give this model a smoke test
// A tiny data set
observe obs(1) = 2;
observe obs(2) = 2;
// Should be solvable exactly
infer posterior;
// Do we get plausible values?
sample list(intercept, slope, line(3))
[1.255954212809891, 0.8266148895039006, 3.735798881321593]
Note that we are using rejection sampling here (infer posterior).
This is fine for this example, but as explored previously, rejection
sampling can become very slow as you increase the difficulty of the
problem (such as by adding more data points, as we will do shortly).
Exercise: For now, evaluate the model's performance on the two data points.
Gather and save a dataset of independent posterior samples of the
parameters and a prediction at some value.
[Hint: You can pass multiple arguments to collect]
(show answer)
Answer:
define data = run(accumulate_dataset(100,
do(posterior,
collect(intercept, slope, line(3)))))
<venture.engine.inference.Dataset object at 0x2b5da88cf390>
Predictive accuracy: Where do you expect the predictive mean to be,
based on the observations we trained the model on? How widely do
you expect it be dispersed, based on the noise in the model? Plot
the predictions: does the data agree with your intuition?
(show answer)
Answer: 2, pretty broad,
plot("h2", data)
[]
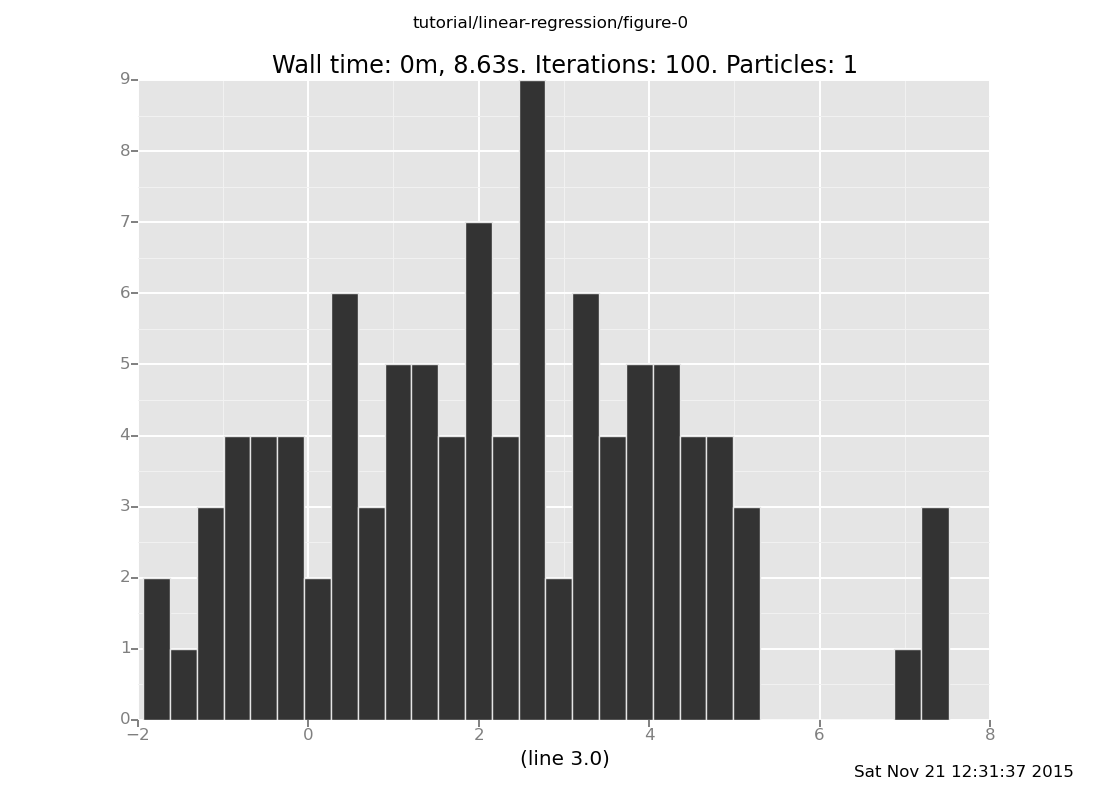
Parameter inference
a. Where do you expect the intercept to be located, and how
concentrated do you expect it to be? Plot the intercept and
check your intuition.
(show answer)
Answer: 2, broad,
plot("h0", data)
[]
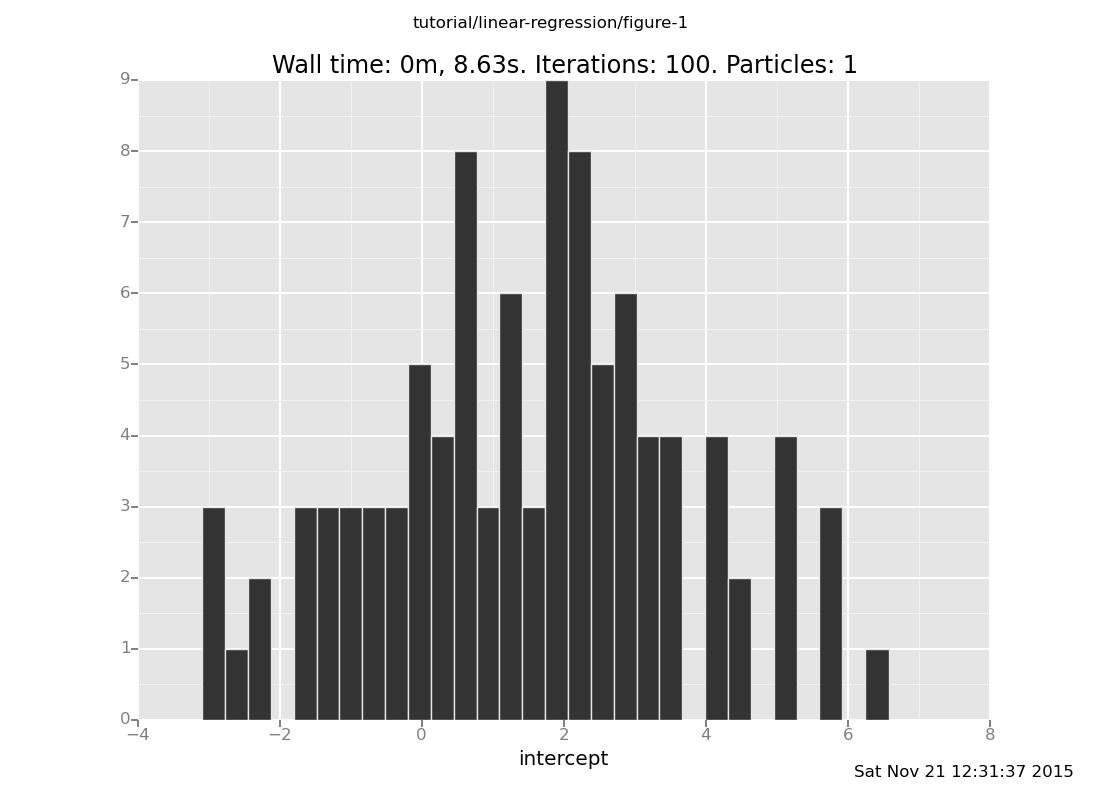
b. Where do you expect the slope to be located, and how
concentrated do you expect it to be? Plot the slope and check
your intuition.
(show answer)
Answer: 0, broad,
plot("h1", data)
[]
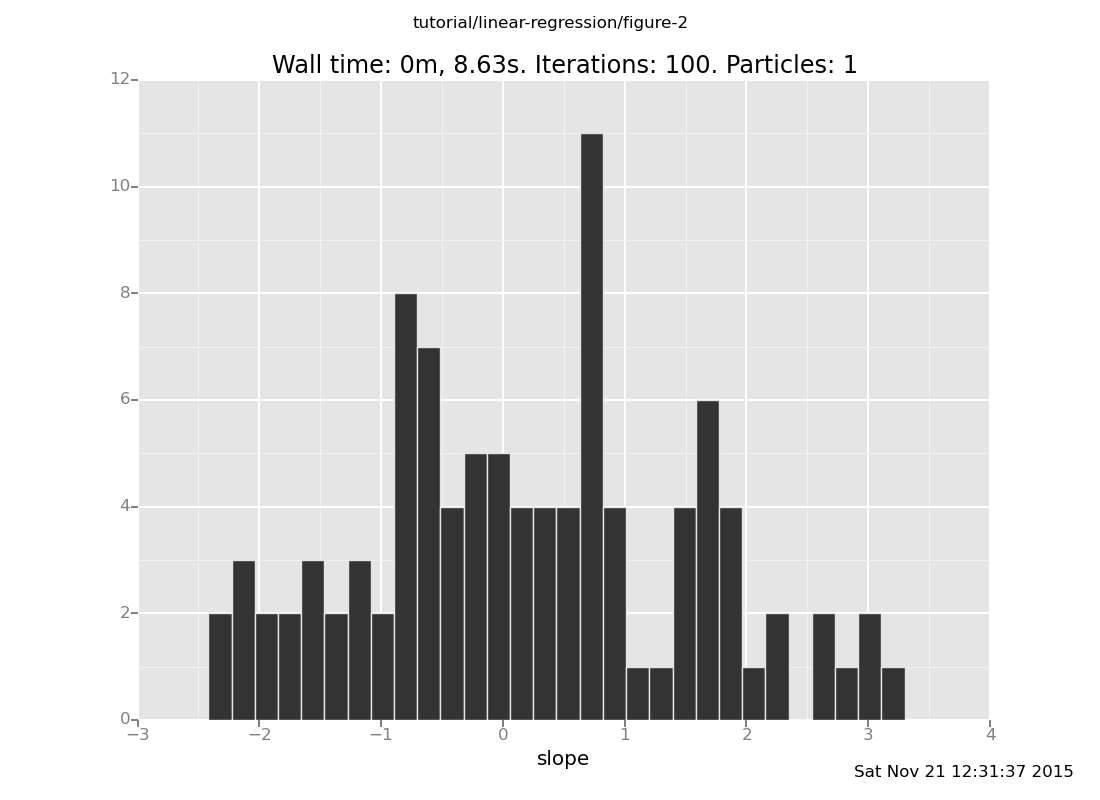
c. What do you expect the relationship between the slope and
intercept to be? Plot them against each other and check your
intuition. Hint: The usual way to get a scatter plot is
p0d1d: p for point and d for direct as opposed to
log scale. A gotcha: if you leave the middle d off, the 0
and 1 will run together into a single index.
(show answer)
Answer: The slope and intercept are anticorrelated, because the
line is trying to pass through around (1.5, 2).
plot("p0d1d", data)
[]
Topic: Working with Datasets
In the previous segment, we started with the simple linear regression
model in lin-reg-1.vnts
$ venture -f lin-reg-1.vnts --interactive
In this segment, we will try it on more data.
There is a little synthetic regression dataset in the file
reg-test.csv. Download it and have a look.
You could, of course, type all this data into your Venture session
as
observe obs(this) = that;
observe obs(that) = the other;
...
but that's pretty tedious. Being a probabilistic programming
language, VentureScript doesn't replicate standard programming facilities
like reading data files, and instead offers close integration with
Python. So the way to do a job like this is to use
pyexec:
run(pyexec("import pandas as pd"));
run(pyexec("frame = pd.read_csv('reg-test.csv')"));
run(pyexec("ripl.observe_dataset('obs', frame[['x','y']].values)"))
[]
list_directives
29: [assume intercept (normal 0 10)]: -0.139405186619
30: [assume slope (normal 0 10)]: 0.720635670215
31: [assume line (lambda (x) (add (mul slope x) intercept))]: <procedure>
32: [assume obs (lambda (x) (normal (line x) 1))]: <procedure>
33: [observe (obs 1) 2]
34: [observe (obs 2) 2]
345: [observe (obs (quote -10.0)) -1.41388873273]
346: [observe (obs (quote -9.0)) -0.543954732733]
347: [observe (obs (quote -8.0)) -0.987143191624]
348: [observe (obs (quote -7.0)) -0.868472949663]
349: [observe (obs (quote -6.0)) -0.0447906285123]
350: [observe (obs (quote -5.0)) 0.701216243962]
351: [observe (obs (quote -4.0)) 0.714336973102]
352: [observe (obs (quote -3.0)) 0.814145307735]
353: [observe (obs (quote -2.0)) 1.85637213834]
354: [observe (obs (quote -1.0)) 1.96607106754]
355: [observe (obs (quote 0.0)) 2.62490949726]
356: [observe (obs (quote 1.0)) 2.53546657413]
357: [observe (obs (quote 2.0)) 3.53349494166]
358: [observe (obs (quote 3.0)) 3.61963267361]
359: [observe (obs (quote 4.0)) 3.70743497539]
360: [observe (obs (quote 5.0)) 4.46624495602]
361: [observe (obs (quote 6.0)) 4.89031624259]
362: [observe (obs (quote 7.0)) 4.94262548685]
363: [observe (obs (quote 8.0)) 5.18359956924]
364: [observe (obs (quote 9.0)) 5.94988043204]
365: [observe (obs (quote 10.0)) 6.22337252036]
If you want to know what just happened there, pyexec is an action of
the inference language that executes the given string of Python code.
The binding environment persists across calls, and begins with the variable ripl bound to the
current session's
Read-Infer-Predict Layer,
which serves as the entry point for using VentureScript as a Python library.
In this case, we used the escape to load our csv file up into a Pandas DataFrame,
and then used the RIPL's observe_dataset method to load it into VentureScript
as individual observations.
OK, so we loaded the data. We could do some inference
infer default_markov_chain(30)
[]
and ask for a prediction at a new x-value
sample line(11)
10.2604776106
but without a look at the data, we don't even know what to expect.
There are of course plenty of ways to plot a dataset; in this case, we
will use VentureScript's plot:
run(pyexec("import venture.lite.value as vv"));
define frame = run(pyeval("vv.VentureForeignBlob(frame[['x', 'y']])"));
plot("p0d1d", frame)
[]
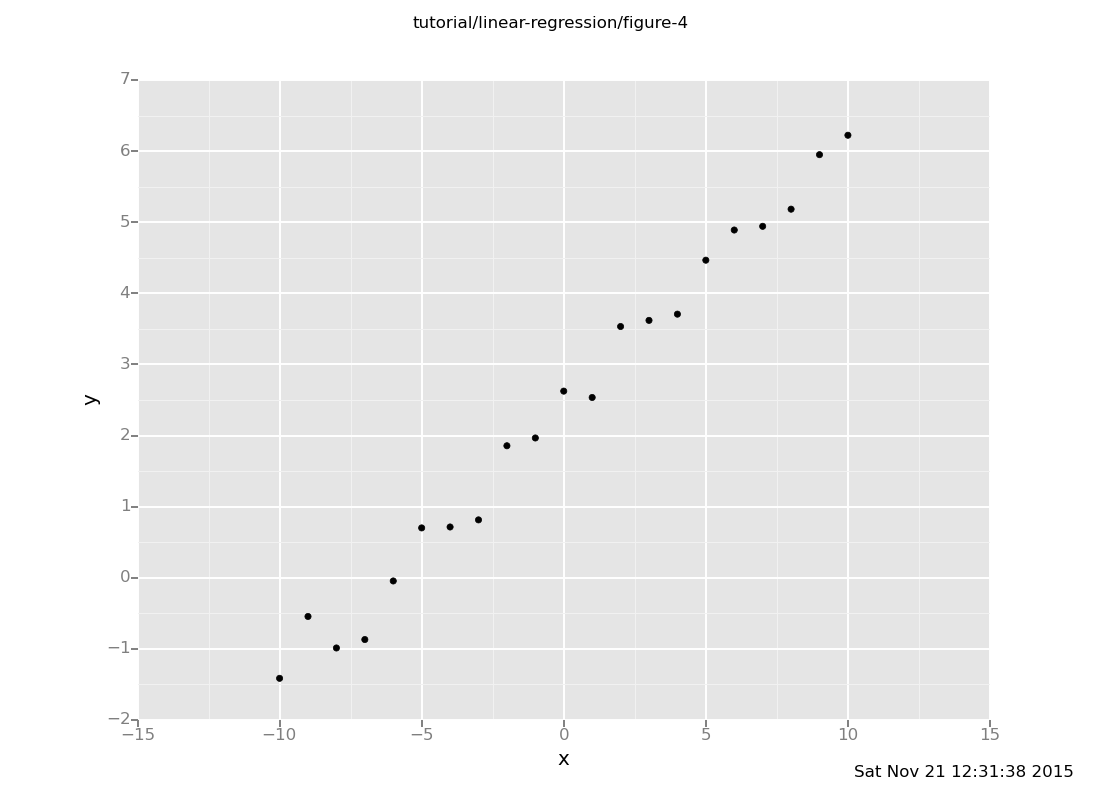
The dataset exhibits a clear linear relationship, and we should expect
line(11) to be around 7.
Let's try using Markov chain inference to fit our linear model. Start
100 independent chains, evolve them for 30 iterations, and record where they end up:
define data = run(accumulate_dataset(100,
do(reset_to_prior,
default_markov_chain(30),
collect(intercept, slope, line(11)))))
<venture.engine.inference.Dataset object at 0x2b5dafbd27d0>
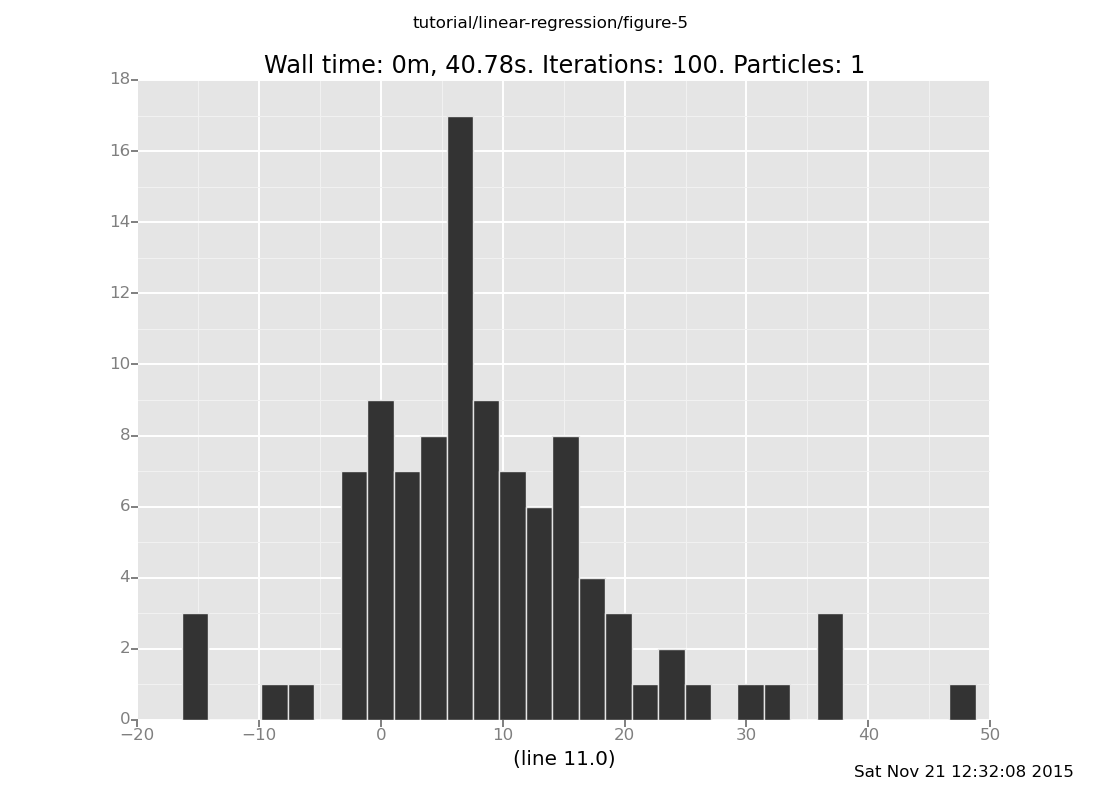
Now we can plot a histogram of the predicted line(11) value.
plot("h2", data)
[]
Topic: Understanding a model with visualization
Is our fit any good? Although we can get some idea of what's going on
by plotting the predictions for a given x, it's much more effective
to be able to see the fitted line plotted alongside the data, and see
what happens over the course of inference.
In order to do so, we can make use of Venture's plugin infrastructure
to load external code written in Python. In this case, we've provided
a custom plugin that produces an animated curve-fitting visualization
that can be updated during the inference program. Download
curve-fitting.py and load it into VentureScript using
run(load_plugin("curve-fitting.py"))
[]
Alternatively, the plugin can be loaded when you launch Venture by
supplying -L curve-fitting.py on the command line.
When the plugin loads, a new window should pop up where the animation
will appear. The plugin provides a callback that can be invoked to
draw a new frame
run(call_back(draw))
[]
which should render a plot that looks like this:
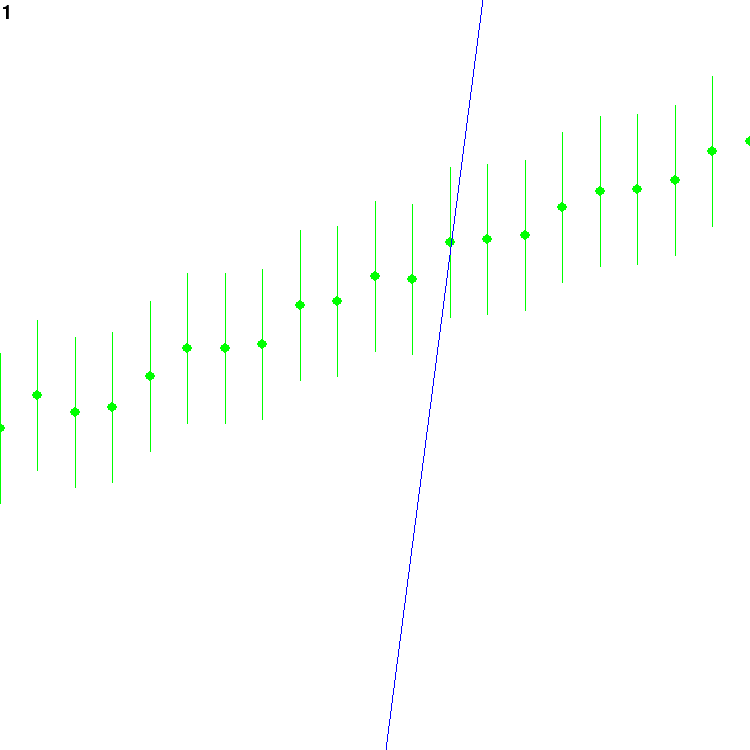
The blue line is the current guess. The green points are the
observations; the error bars around them reach twice the standard
deviation of the noise, above and below. The number at the top left
is the count of how many frames have been drawn.
We can also plot multiple frames with inference commands interleaved
infer reset_to_prior;
infer repeat(30, do(default_markov_chain(1),
call_back(draw)))
[]
The repeat command works
similarly to accumulate_dataset, but in this case we aren't
concerned with collecting the data since it's being rendered in the
visualization instead. Together, this command will run the Markov
chain for 30 iterations, drawing an animation frame after each
iteration.
Another useful command which works well with visualization is
loop, which runs the given
action continually in the background. You can use it like this
infer loop(do(default_markov_chain(5),
call_back(draw)))
None
which will iterate the Markov chain and draw an animation frame every
5 iterations, continuing to do so indefinitely. You can turn it off
with
stop_continuous_inference
Exercise: Play with this visualization to see how inference is doing.
Use the loop command to run the Markov chain continually and draw
an animation frame after every iteration. Watch the animation while
it runs and confirm that the line eventually approaches a good fit
to the data. Does it take gradual steps or sudden jumps?
(show answer)
Answer: It takes sudden jumps, with varying amount of time between
jumps.
infer loop(do(default_markov_chain(1), call_back(draw)))
None
How many iterations does it take for the chain to converge? Modify
the inference loop to call reset_to_prior after every 30
iterations of the Markov chain. (Hint: You may want to use repeat
and do.) Is the Markov chain able to find a good fit within 30
iterations? What about 100 iterations? Try playing with the number
of iterations to see how many it takes to reliably find a
reasonable fit.
(show answer)
Answer: 30 is not enough, and it may not even be obvious visually
when it's resetting to the prior. 100 is better, but still often
fails to find a fit; 200-300 is clearly able to find it most of the
time.
infer loop(do(reset_to_prior,
repeat(30, do(default_markov_chain(1),
call_back(draw)))))
None
(If you're still running the infer loop, turn it off for this
part.)
stop_continuous_inference
We can use resample to run multiple independent chains and plot
all of the resulting lines in the same window, to get some sense of
the posterior distribution over lines. Resample 20 particles
(remember to reset them to the prior), run them for 30 iterations,
and look at the resulting animation. Does it look like the
distribution of lines converges to something plausible? If you're
impatient, try increasing the number of iterations as above.
(show answer)
Answer: Some of the samples do concentrate around the correct line,
but again 30 iterations is not enough to converge.
infer resample(20);
infer do(reset_to_prior,
repeat(30, do(default_markov_chain(1),
call_back(draw))));
infer resample(1);
[]
Inference alternatives
a. In the optional subunit at the end of the previous
chapter,
we saw how to construct our own Markov chains that used custom
transition operators, with a drift kernel example. Experiment
with using a drift kernel for this problem. How quickly can you
get it to converge by choosing a good proposal distribution?
(show answer)
Here is an example program using a Cauchy proposal kernel (you
may have used something different):
```
define driftkernel = proc(x) { studentt(1, x, 1) };
define driftmarkovchain =
mhcorrect(
onsubproblem(default, all,
symmetriclocalproposal(drift_kernel)));
infer do(resettoprior,
repeat(30, do(driftmarkovchain,
call_back(draw))))
```
[]
The Cauchy distribution has heavy tails, which means that this
will usually take small steps but occasionally propose large
steps. This seems to work fairly well here.
b. As an alternative to Markov chains, we can use gradient ascent
to find the best fit. Venture provides an inference primitive,
map (for "maximum a
posteriori") which deterministically climbs the gradient toward
the most probable parameter values. It can be invoked as
follows:
infer repeat(30, do(map(default, all, 0.001, 1, 1),
call_back(draw)))
[]
This has a very different character than the inference
primitives we've seen so far. Experiment with it, tweaking the
step size (the "0.001"; be careful setting it too high!) and
using reset_to_prior to start from a few different initial
conditions. Can you think of some advantages and disadvantages
of gradient methods compared to Monte Carlo methods?
(show answer)
Answer: Gradient ascent can move toward high-probability
regions of the parameter space faster and more directly than
Monte Carlo methods. However, the convergence of gradient
ascent highly depends on the shape of the posterior probability
density function; if the posterior density is too flat, it will
move very slowly, whereas if the posterior density is too steep
relative to the step size, it may overshoot and spiral off to
infinity. Additionally, if there are local maxima, gradient
ascent will not be able to escape them. (Markov chains are also
susceptible to local maxima, although less so.)
Convergence aside, an important conceptual difference is that
gradient ascent gives only a point estimate of the single most
probable values. In contrast, sampling methods return values
that are (approximately) distributed according to the full
posterior distribution, allowing the uncertainty in those
values to be quantified and taken into account when making
decisions.
Elaboration: Outlier Detection
So far, we haven't really done anything beyond what standard tools for
linear least squares regression can do. Now we will look at what
happens if the data may contain outliers: points that do not fit in
with the rest. In ordinary least squares regression, outliers can
disproportionally influence the result and so are often excluded
before running the regression. Here, we will take a different approach
by extending our probabilistic model to take outliers into account.
clear
We start with our linear model much the same as before
run(load_plugin("curve-fitting.py"));
assume intercept = normal(0,7);
assume slope = normal(0,4);
assume line = proc(x) { slope * x + intercept }
<procedure>
but this time, each point has some probability of being an outlier.
assume outlier_prob = uniform_continuous(0.01,0.3);
assume is_outlier = mem(proc(i) { flip(outlier_prob) })
<procedure>
(Here we use mem to create an
indexed set of random variables is_outlier(i) for each i, which
indicate whether the i-th point is an outlier.)
What does it mean for a point to be an outlier? Unlike most of the
other points, which we can expect to be near the value predicted by
our linear equation, an outlier could just as likely be anywhere. We
will model that by saying that for an outlier point, the y-value is
drawn from normal(0,20).
assume obs_outlier = proc(x) { normal(0,20) }
<procedure>
Non-outlier points, as before, are centered on the line but subject to
some Gaussian noise. This time, rather than assuming a fixed amount of
noise, we will put a prior over the amount of noise as well and see
how well we can infer it:
assume noise = gamma(1,1);
assume obs_inlier = proc(x) { normal(line(x), noise) }
<procedure>
Putting that all together, our full data model is
assume obs = proc(i,x) {
if (is_outlier(i)) {
obs_outlier(x)
} else {
obs_inlier(x)
}
}
<procedure>
Whew. If you don't want to type all that in, it's also in
lin-reg-2.vnts.
Let's try this with some data
observe obs(0,1) = 2.5;
observe obs(1,4) = 1;
observe obs(2,-3) = -1;
observe obs(3,-5) = 6
and load it into our visualization
run(load_plugin("curve-fitting.py"));
infer loop(do(default_markov_chain(1),
call_back(draw)))
None
This should spend a while considering different hypotheses and whether
or not to treat each point as an outlier. Eventually, it might find
this configuration and settle on it for a while:
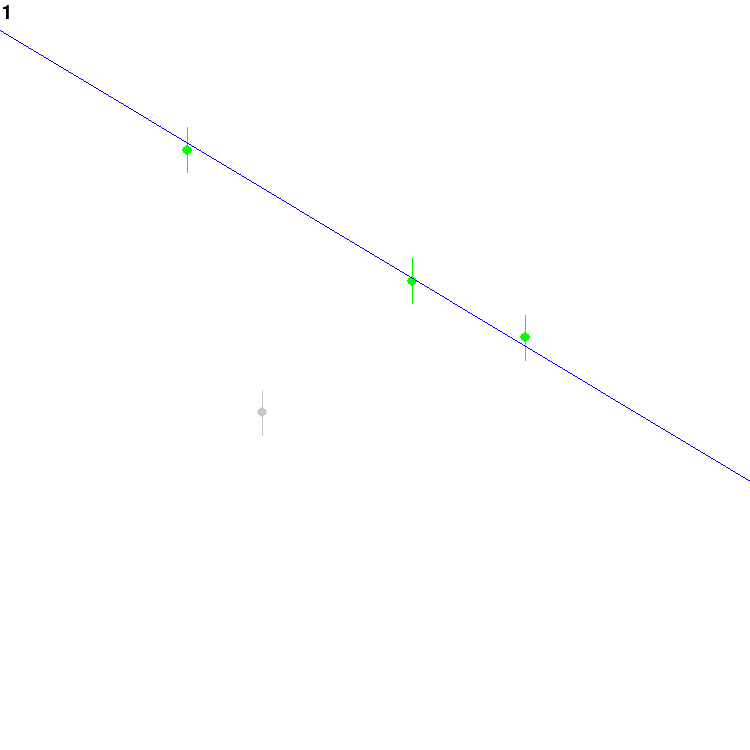
The grey dot is currently assumed to be an outlier, whereas the green
ones are inliers.
stop_continuous_inference
If you are used to least squares regression, the above model may look
a bit weird. A search process that was trying to minimize, say,
squared error on inliers would be very happy to just call every data
point an outlier and be done with it. Why doesn't the Markov chain
decide to do that?
This is an instance of the phenomenon known as Bayesian Occam's
Razor. On this example, it works as follows: Since we are being
fully Bayesian here, we have to give a probability model for the
outliers as well as the inliers. Since it's a probability model, its
total mass per data point must add up to 1, just like the inlier
model's total mass per data point. Since the point of the outlier
model is to assign reasonable probability to a broad range of points
(outliers!), it perforce cannot assign very high mass to any of them
-- necessarily lower than the mass the inlier model assigns to points
sufficiently near the line. Consequently, several nearly colinear
points are much more likely to be inliers on a line than to all be
outliers; and the Markov chain, being a correct algorithm, finds this.
Philosophically, one might call the outlier model "complex", in the
sense of being high entropy; and Bayesian Occam's Razor automatically
causes the posterior to prefer lower entropy -- "simpler" --
explanations.
Now that we have a more complex model with more moving parts, we'd
like to be able to treat each of those parts independently when it
comes to deciding how to do inference. VentureScript provides a mechanism to
tag different parts of the model program and then refer to those tags
from inference commands.
Let's see how this works. We'll load the same model as before, but
this time we'll tag each of the random choices made in the model so
that we can refer to them separately (lin-reg-3.vnts):
clear
run(load_plugin("curve-fitting.py"));
assume intercept = tag("param", "intercept", normal(0,7));
assume slope = tag("param", "slope", normal(0,4));
assume line = proc(x) { slope * x + intercept };
assume noise = tag("hyper", "noise", gamma(1, 1));
assume outlier_prob = tag("hyper", "outlier_prob", uniform_continuous(0.01, 0.3));
assume is_outlier = mem(proc(i) { tag("outlier", i, flip(outlier_prob)) });
assume obs = proc(i, x) {
if (is_outlier(i)) {
normal(0,20)
} else {
normal(line(x), noise)
}
};
<procedure>
Each tag command takes three arguments: a scope identifier, a block
identifier, and the expression it applies to. Scope and block
identifiers can be any unique expression. Inference commands can then
use these identifiers to target a particular scope and block. For
example, we can do Markov chain inference on just the line parameters
using
infer mh("param", one, 1);
[]
The mh inference command is the same as default_markov_chain,
except that it allows you to specify a scope and block to operate on;
in fact, default_markov_chain(x) is an alias for mh(default, one,
x). Here default is a keyword that refers to the default scope
containing all random choices in the model, and one means to pick
one randomly from the blocks associated with the scope.
What are scoped inference commands good for? Here are some use cases:
Specifying how to divide effort between different parts of the
model. For instance, one issue with using the default Markov chain
on the model with outliers is that the default chain will consider
each random choice in the model equally often, but the model
contains one random choice for every data point that determines
whether it's an outlier or not, so as the number of data points
increases it will spend most of its time categorizing points as
outliers and relatively little time actually fitting the line
parameters. Using scopes, we can explicitly decide how many
iterations to allocate to outlier detection to avoid having it take
a disproportionate amount of effort.
Using different inference strategies for different parts of the
model. For instance, we may want to use enumerative Gibbs for the outlier detection, and
drift proposals for the line parameters. This is especially useful
when some inference strategies only make sense for some types of
variables; in this case, enumerative Gibbs is only possible for
discrete variables and normal drift proposals only make sense for
continuous variables.
Let's apply these ideas to our model. Load the observations again
observe obs(0,1) = 2.5;
observe obs(1,4) = 1;
observe obs(2,-3) = -1;
observe obs(3,-5) = 6
and run the following inference program:
define drift_kernel = proc(x) { student_t(1, x, 1) };
define drift_mh =
proc(scope, block, transitions) {
repeat(transitions,
mh_correct(
on_subproblem(scope, block,
symmetric_local_proposal(drift_kernel))))
};
infer loop(do(drift_mh("param", all, 2),
mh("hyper", one, 1),
gibbs("outlier", one, 1),
call_back(draw)))
None
This will alternate between the different tagged parts of the model,
using a different inference strategy for each one.
Exercise: Compare this to the infer loop that only used
default_markov_chain from earlier. What qualitative differences in
behavior do you see? Try adjusting the number of iterations allocated
to each scope and see what happens.
(show answer)
Answer: The line moves more often, because the program spends less
time on uselessly reconsidering whether points are outliers and more
time on proposing new line parameters.
Giving more iterations to the line parameters (say, mh("param",
one, 5)) will cause the line to move even more, but it may miss the
opportunity to reduce the noise level or flag a point as an outlier
when it doesn't fit.
Giving more iterations to the noise hyperparameter or the outlier
detection will make these opportunities more likely, but may also
waste time reconsidering the same random choices over and over just
like default_markov_chain does.
Exercise: What happens if you replace drift_mh with map (gradient
ascent)? Hint: Make sure to pass the appropriate scope and block to
map.
(show answer)
The line now moves smoothly toward the points that are marked as
non-outliers. On the flip side, it is no longer able to explore
alternative hypothesis once it has settled on a (possibly suboptimal)
fit.
infer loop(do(map("param", all, 0.001, 10, 1),
mh("hyper", one, 1),
gibbs("outlier", one, 1),
call_back(draw)))
None
stop_continuous_inference







